Density-based intrinsic persistent homology & applications to time series analysis
XIMENA FERNANDEZ
Durham University
UK Centre for Topological Data Analysis
Applied CATS Seminar
KTH Royal Institute of Technology - 28th February 2023
Density-based
intrinsic persistent homology
$\bullet$ Fernandez X., Borghini E., Mindlin G. & Groisman P. Intrinsic persistent homology via density-based metric learning, Journal of Machine Learning Research, 2023 (to appear) arXiv:2012.07621
Homology inference
Let $\mathbb{X}_n = \{x_1,...,x_n\}\subseteq \mathbb{R}^D$ be a finite sample.

Homology inference
Let $\mathbb{X}_n = \{x_1,...,x_n\}\subseteq \mathbb{R}^D$ be a finite sample.
Assume that:
- $\mathbb{X}_n$ is a sample of a compact manifold $\mathcal M$ of dimension $d$.
- The points are sampled according to a density $f\colon \mathcal M\to \mathbb R$.
Goal: Infer $H_\bullet(\mathcal M)$

Homology inference
Metric space: $(\mathbb X_n, d_E)\sim (\mathcal M, d_E)$
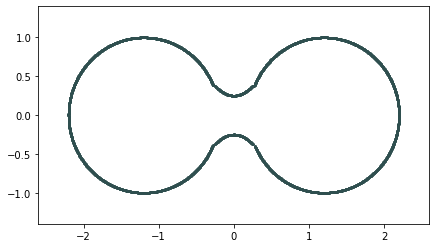
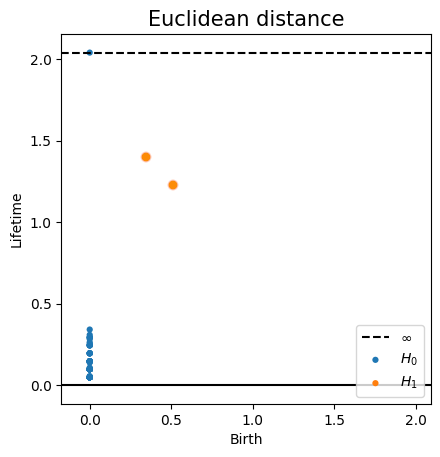
$\bullet ~ ~\mathrm{Rips}_\epsilon(\mathcal{M}, d_E)\simeq \mathcal{M}$ for $\epsilon < 2 \sqrt{\frac{D+1}{2D}}\mathrm{rch}(\mathcal{M})~~$ (Kim, Shin, Chazal, Rinaldo & Wasserman, 2020)
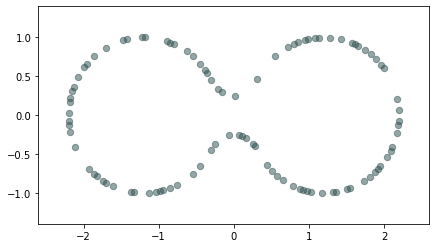
Homology inference
Metric space: $(\mathbb X_n, d_{kNN})\sim (\mathcal M, d_\mathcal{M})~~~$
(Bernstein, De Silva, Langford & Tenenbaum, 2000)

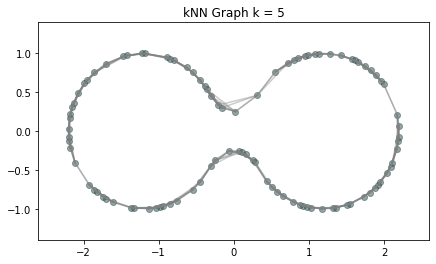
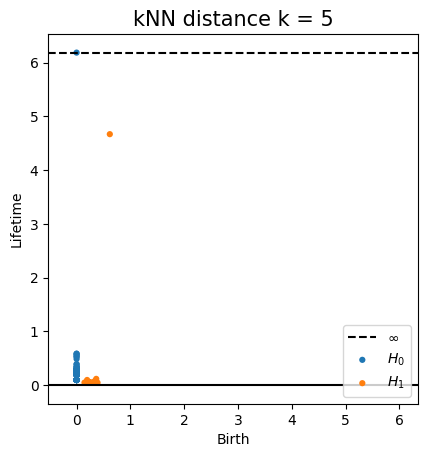
$\bullet ~ ~\mathrm{Rips}_\epsilon(\mathcal{M}, d_\mathcal{M})\simeq \mathcal{M}$ for $\epsilon < \mathrm{conv}(\mathcal{M}, d_{\mathcal{M}})~~$ (Hausmann, 1995; Latschev, 2001)
Homology inference
Metric space: $(\mathbb X_n, d_{kNN})\sim (\mathcal M, d_\mathcal{M})$

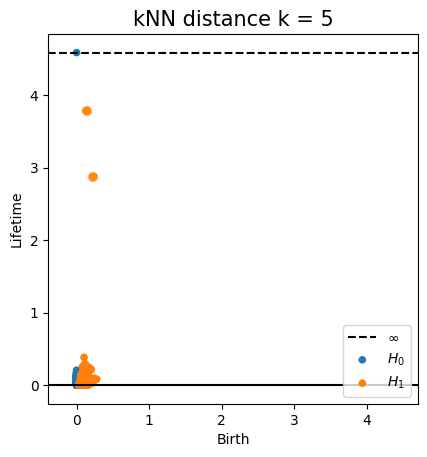
Homology inference
Metric space: $(\mathbb X_n, d_{kNN})\sim (\mathcal M, d_\mathcal{M})$



Homology inference
Metric space: $(\mathbb X_n, d_{kNN})\sim (\mathcal M, d_\mathcal{M})$


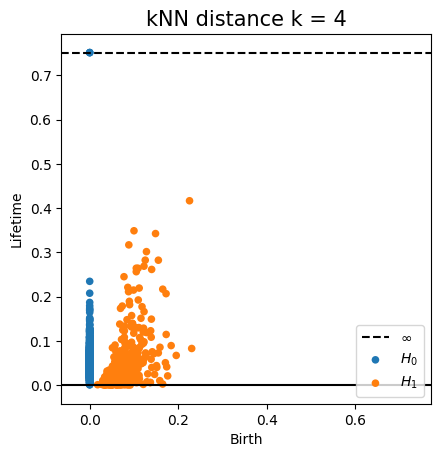
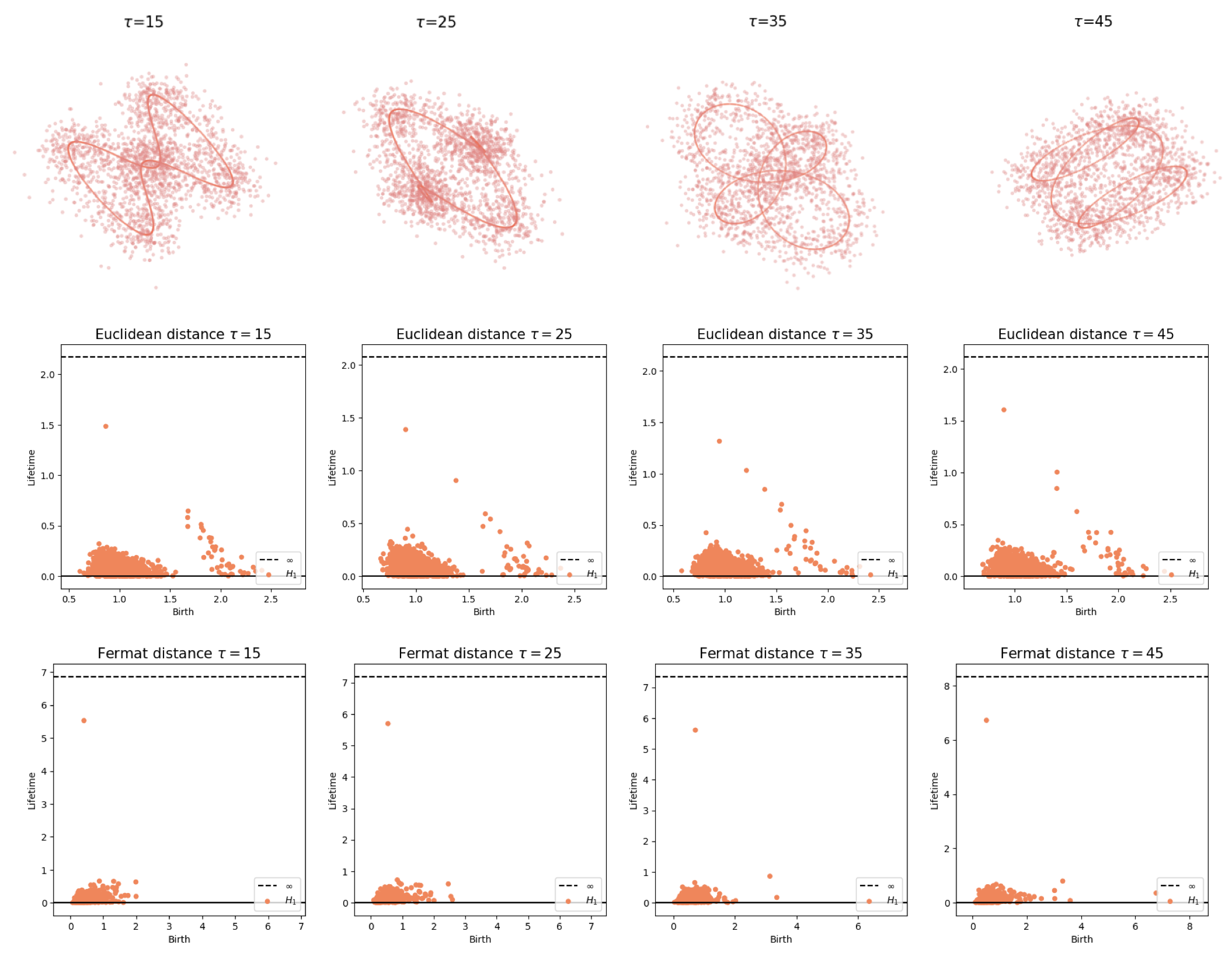
Fermat distance
(Mckenzie & Damelin, 2019) (Groisman, Jonckheere & Sapienza, 2022)Let $\mathbb{X}_n = \{x_1,...,x_n\}\subseteq \mathbb{R}^D$ be a finite sample.
For $p> 1$, the Fermat distance between $x,y\in \mathbb{R}^D$ is defined by \[ d_{\mathbb{X}_n, p}(x,y) = \inf_{\gamma} \sum_{i=0}^{r}|x_{i+1}-x_i|^{p} \] over all paths $\gamma=(x_0, \dots, x_{r+1})$ of finite length with $x_0=x$, $x_{r+1} = y$ and $\{x_1, x_2, \dots, x_{r}\}\subseteq \mathbb{X}_n$.
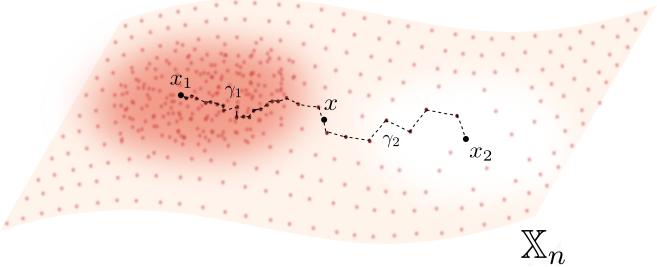
Fermat distance
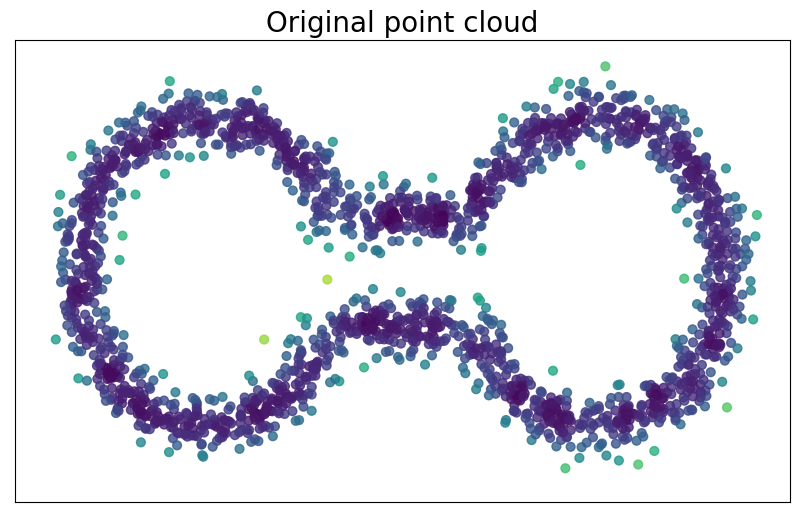
Fermat distance


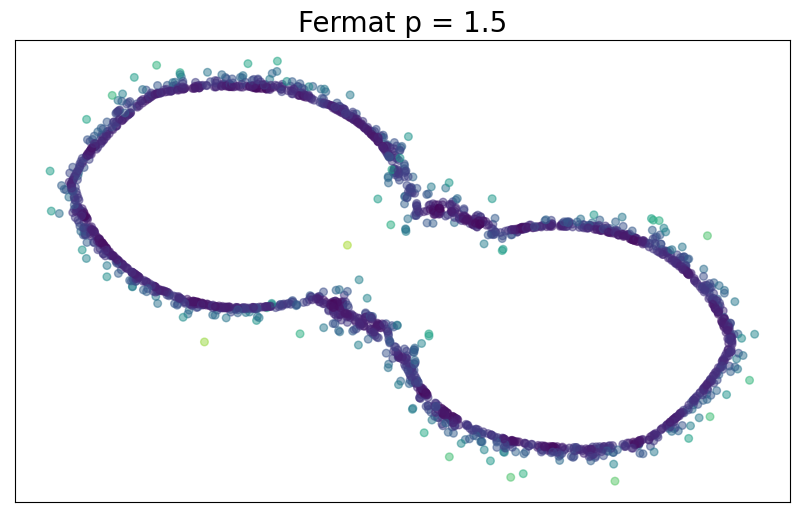
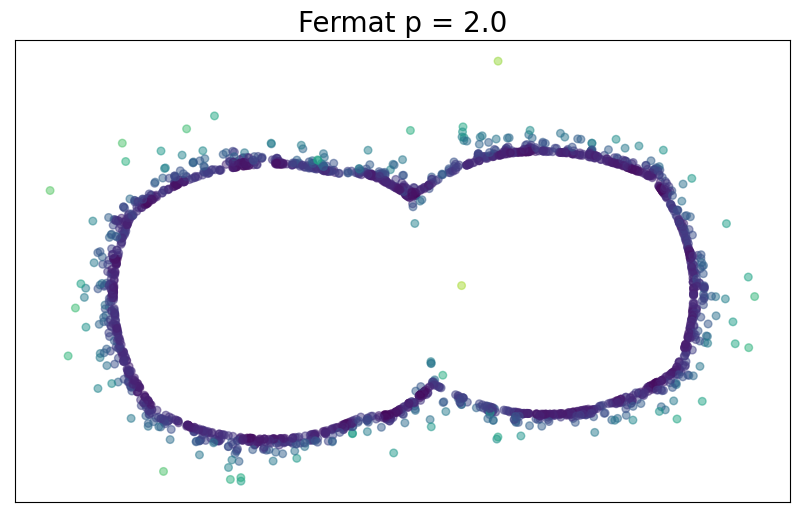
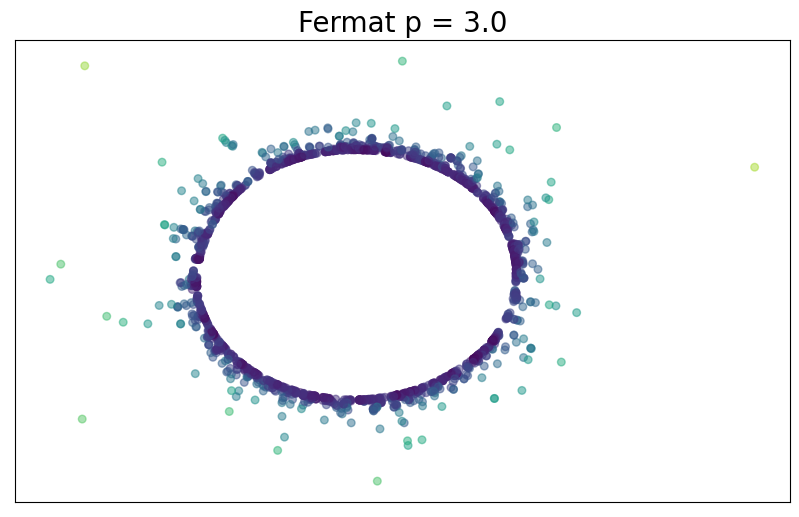
Fermat distance





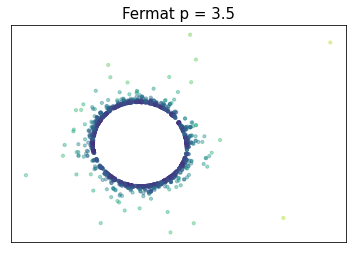
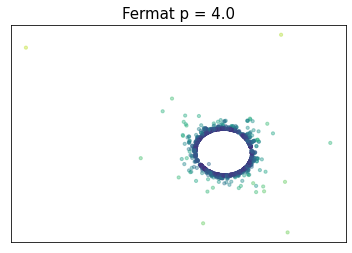
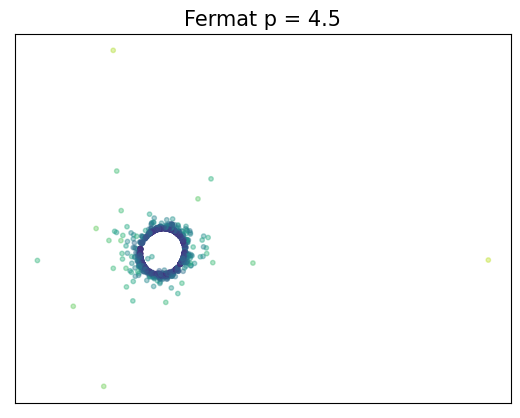
Density-based geometry
(Hwang, Damelin & Hero, 2016)Let $\mathcal M \subseteq \mathbb{R}^D$ be a manifold and let $f\colon\mathcal{M}\to \mathbb{R}_{>0}$ be a smooth density.
For $q>0$, the deformed Riemannian distance* in $\mathcal{M}$ is \[d_{f,q}(x,y) = \inf_{\gamma} \int_{I}\frac{1}{f(\gamma_t)^{q}}||\dot{\gamma}_t|| dt \] over all $\gamma:I\to \mathcal{M}$ with $\gamma(0) = x$ and $\gamma(1)=y$.
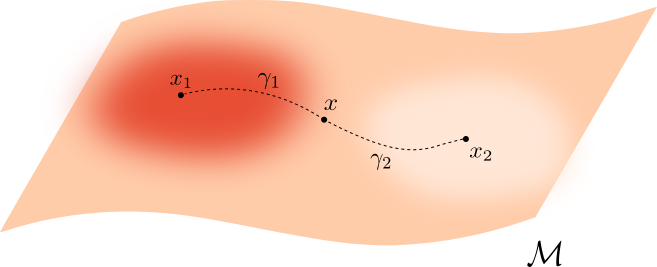
* Here, if $g$ is the inherited Riemannian tensor, then $d_{f,q}$ is the Riemannian distance induced by $g_q= f^{-2q} g$.
Convergence results
(Groisman, Jonckheere & Sapienza, 2022)
\[C(n,p,d) d_{\mathbb{X}_n,p}(x,y)\xrightarrow[n\to \infty]{a.s.}d_{f,q}(x,y) ~~~ \text{ for } q = (p-1)/d\text{ and every }x,y\in \mathcal{M}\]
Convergence results
(Groisman, Jonckheere & Sapienza, 2022)
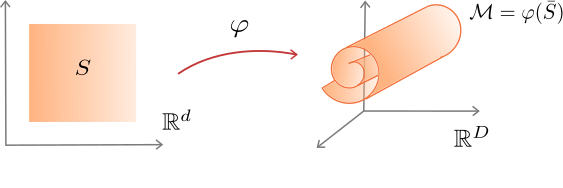
Let $\mathcal{M}$ be an isometric $C^1$ $d$-dimensional manifold embedded in $\mathbb{R}^D$. That is, there exists $S\subseteq \mathbb{R}^d$ an open connected set and $\phi:S \to \mathbb{R}^D$ an isometric transformation such that $\phi(\bar S)=\mathcal{M}. $
\[C(n,p,d) d_{\mathbb{X}_n,p}(x,y)\xrightarrow[n\to \infty]{a.s.}d_{f,q}(x,y) ~~~ \text{ for } q = (p-1)/d\text{ and every }x,y\in \mathcal{M}\]
Convergence results
(Groisman, Jonckheere & Sapienza, 2022)
Let $\mathcal{M}$ be an isometric $C^1$ $d$-dimensional manifold embedded in $\mathbb{R}^D$. That is, there exists $S\subseteq \mathbb{R}^d$ an open connected set and $\phi:S \to \mathbb{R}^D$ an isometric transformation such that $\phi(\bar S)=\mathcal{M}. $

Theorem (Groisman, Jonckheere, Sapienza, 2022)
Given $p>1$ and $q=(p-1)/d$, there exists a constant $\mu = \mu(p,d)$ such that, for any $x,y\in\mathcal{M}$, \[ \lim_{n\to +\infty} n^q d_{\mathbb{X}_n, p}(x,y) = \mu d_{f,q}(x,y )~ \text{almost surely.} \]
Convergence results
(Hwang, Damelin & Hero, 2016)
Let $\mathcal{M}$ be a closed smooth $d$-dimensional manifold embedded in $\mathbb{R}^D$.
\[C(n,p,d) L_{\mathbb{X}_n,p}\underset{n \to \infty}{\overset{a.s}{\rightrightarrows}}d_{f,q}~~~ \text{ for } q = (p-1)/d\text{ in } \{(x,y)\in \mathcal{M}: d_{\mathcal M}(x,y)\geq b\}\]
Convergence results
(Hwang, Damelin & Hero, 2016)
Let $\mathcal{M}$ be a closed smooth $d$-dimensional manifold embedded in $\mathbb{R}^D$.
Consider the length of the power-weighted shortest path \[L_{\mathbb X_n, p}(x,y) := \inf_{\gamma}\sum_{i=0}^{k}d_{\mathcal M}(x_{i+1},x_i)^{p}\] over all paths $\gamma=(x_0, \dots, x_{r+1})$ of finite length with $x_0=x$, $x_{r+1} = y$ and $\{x_1, \dots, x_r\}\subseteq \mathbb{X}_n$.
\[C(n,p,d) L_{\mathbb{X}_n,p}\underset{n \to \infty}{\overset{a.s}{\rightrightarrows}}d_{f,q} ~~~ \text{ for } q = (p-1)/d\text{ in } \{(x,y)\in \mathcal{M}: d_{\mathcal M}(x,y)\geq b\}\]
Convergence results
(Hwang, Damelin & Hero, 2016)
Let $\mathcal{M}$ be a closed smooth $d$-dimensional manifold embedded in $\mathbb{R}^D$.
Consider the length of the power-weighted shortest path \[L_{\mathbb X_n, p}(x,y) := \inf_{\gamma}\sum_{i=0}^{k}d_{\mathcal M}(x_{i+1},x_i)^{p}\] over all paths $\gamma=(x_0, \dots, x_{r+1})$ of finite length with $x_0=x$, $x_{r+1} = y$ and $\{x_1, \dots, x_r\}\subseteq \mathbb{X}_n$.
Theorem (Hwang, Damelin, Hero, 2016)
Given $\varepsilon > 0$ and $b>0$, there exists constants $\mu = \mu(d,p)>0$ and $\theta = \theta(\varepsilon)>0$ such that, for all sufficiently large $n$, \[ \mathbb {P} \left(\sup_{\substack{x,y: d_{\mathcal M}(x,y)\geq b}}\left|\frac{n^{q}L_{\mathbb X_n, p}(x,y)}{ \mu d_{f,q}(x,y)}-1\right|>\varepsilon\right)\leq \exp(-\theta n^{1/(d+2p)}) \] In particular, for every $x,y\in \mathcal M$, $ \lim_{n\to +\infty} n^{(p-1)/d} L_{\mathbb X_n, p}(x,y) = \mu d_{f,p}(x,y )~ \text{almost surely.} $
Convergence results
(F., Borghini, Mindlin & Groisman, 2023)
Let $\mathcal{M}$ be a closed smooth $d$-dimensional manifold embedded in $\mathbb{R}^D$.
\[\big(\mathbb{X}_n, C(n,p,d) d_{\mathbb{X}_n,p}\big)\xrightarrow[n\to \infty]{GH}\big(\mathcal{M}, d_{f,q}\big) ~~~ \text{ for } q = (p-1)/d\]
Theorem (F., Borghini, Mindlin, Groisman, 2023)
Given $p>1$ and $q=(p-1)/d$, there exists a constant $\mu = \mu(p,d)$ such that for every $\lambda \in \big((p-1)/pd, 1/d\big)$ and $\varepsilon>0$ there exist $\theta>0$ satisfying \[ \mathbb{P}\left( d_{GH}\left(\big(\mathcal{M}, d_{f,q}\big), \big(\mathbb{X}_n, {\scriptstyle \frac{n^{q}}{\mu}} d_{\mathbb{X}_n, p}\big)\right) > \varepsilon \right) \leq \exp{\left(-\theta n^{(1 - \lambda d) /(d+2p)}\right)} \] for $n$ large enough.
Convergence of persistence diagrams
\[\big(\mathbb{X}_n, C(n,p,d) d_{\mathbb{X}_n,p})\big)\xrightarrow[n\to \infty]{GH}\big(\mathcal{M}, d_{f,q}\big) ~~~ \text{ for } q = (p-1)/d\]
+Stability \[d_B\Big( \mathrm{dgm}\big(\mathrm{Filt}(\mathbb X_n, C(n,p,d) d_{\mathbb{X}_n,p})\big), \mathrm{dgm}\big(\mathrm{Filt}(\mathcal M, d_{f,q})\big)\Big)\leq 2 d_{GH}\big((\mathbb X_n,C(n,p,d) d_{\mathbb{X}_n,p}),(\mathcal M,d_{f,q})\big)\]
Convergence of persistence diagrams
\[\big(\mathbb{X}_n, C(n,p,d) d_{\mathbb{X}_n,p})\big)\xrightarrow[n\to \infty]{GH}\big(\mathcal{M}, d_{f,q}\big) ~~~ \text{ for } q = (p-1)/d\]
+Stability \[d_B\Big( \mathrm{dgm}\big(\mathrm{Filt}(\mathbb X_n, C(n,p,d) d_{\mathbb{X}_n,p})\big), \mathrm{dgm}\big(\mathrm{Filt}(\mathcal M, d_{f,q})\big)\Big)\leq 2 d_{GH}\big((\mathbb X_n,C(n,p,d) d_{\mathbb{X}_n,p}),(\mathcal M,d_{f,q})\big)\]
$\Downarrow$\[\mathrm{dgm}(\mathrm{Filt}(\mathbb{X}_n, {C(n,p,d)} d_{\mathbb{X}_n,p}))\xrightarrow[n\to \infty]{B}\mathrm{dgm}(\mathrm{Filt}(\mathcal{M}, d_{f,q})) ~~~ \text{ for } q = (p-1)/d\]
Fermat-based persistence diagrams

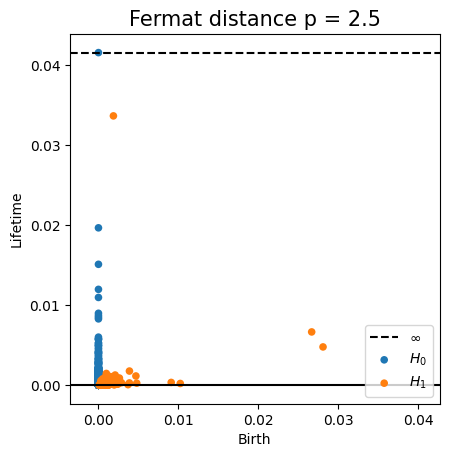
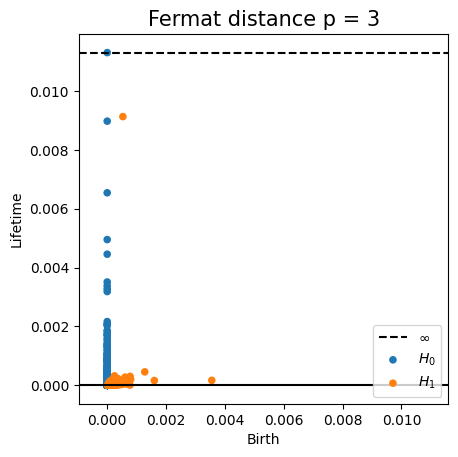
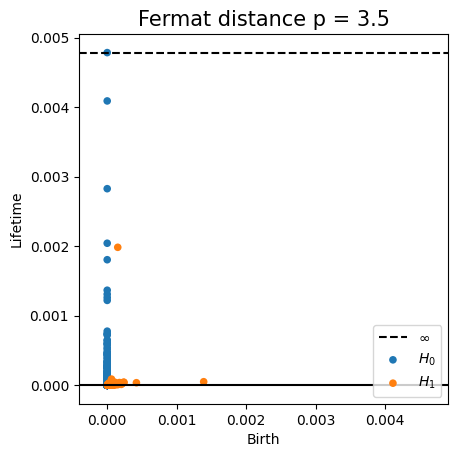
Fermat-based persistence diagrams


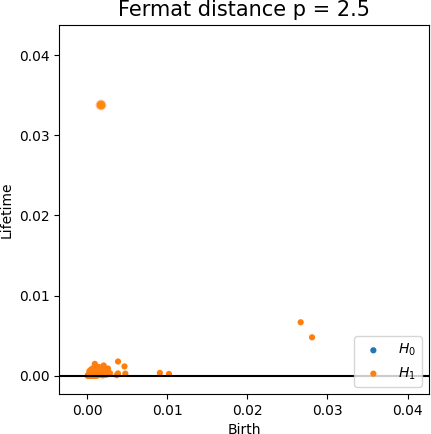
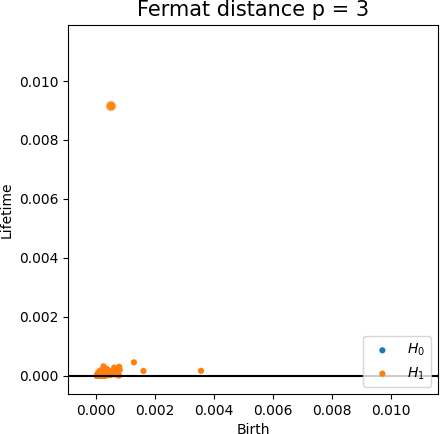
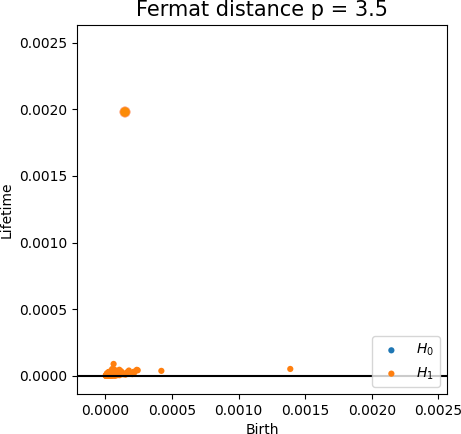
Fermat-based persistence diagrams


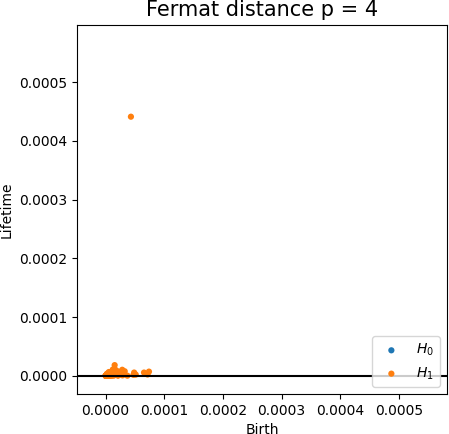
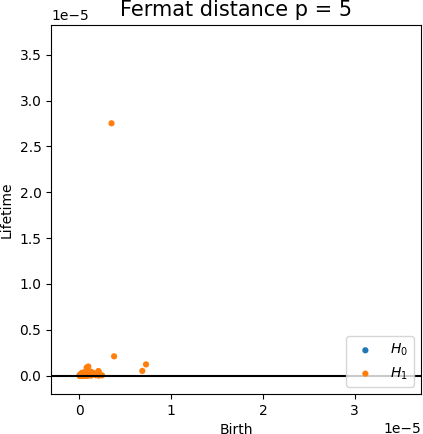
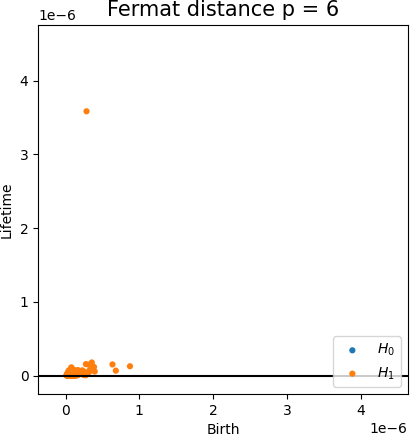
Fermat-based persistence diagrams
Intrinsic reconstruction





Fermat-based persistence diagrams
Robustness to outliers
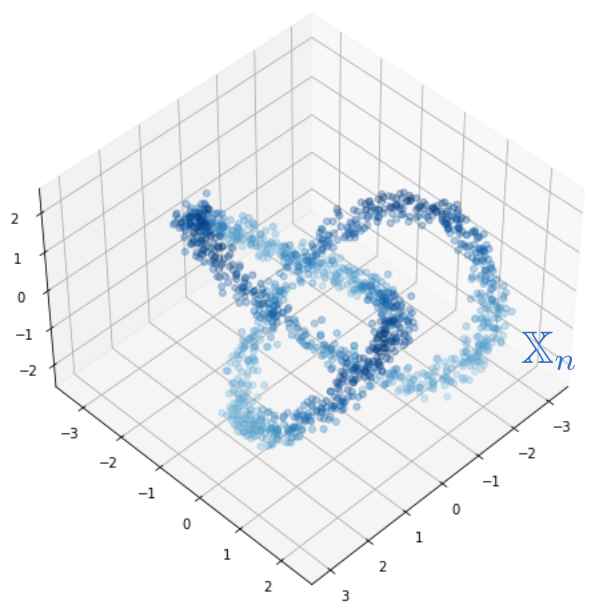
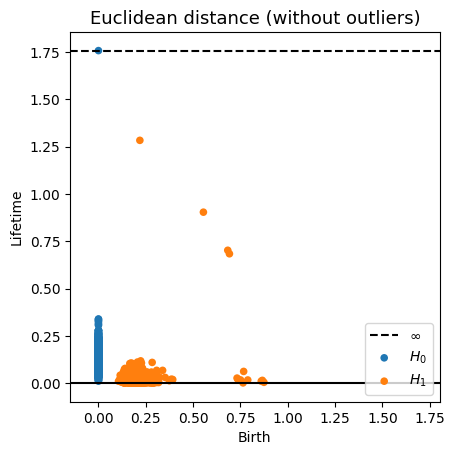
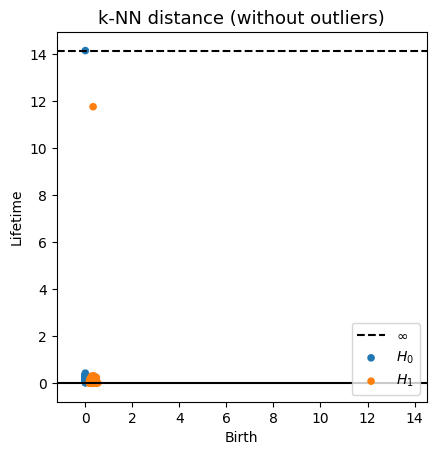
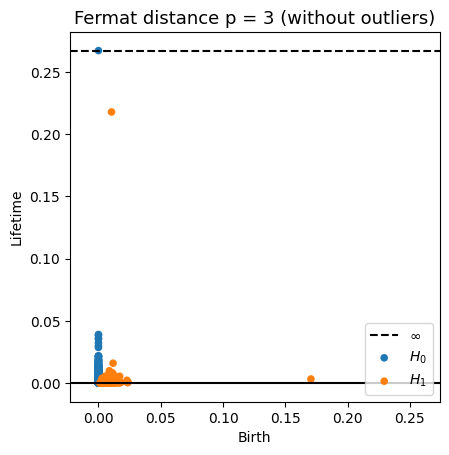
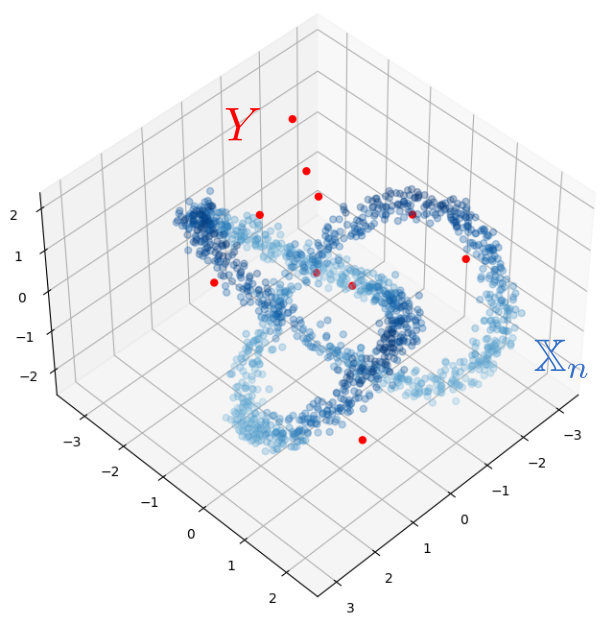
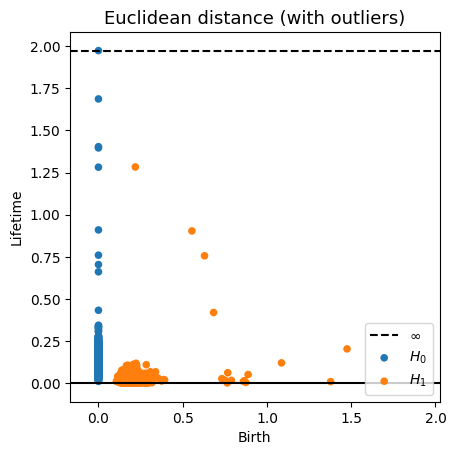
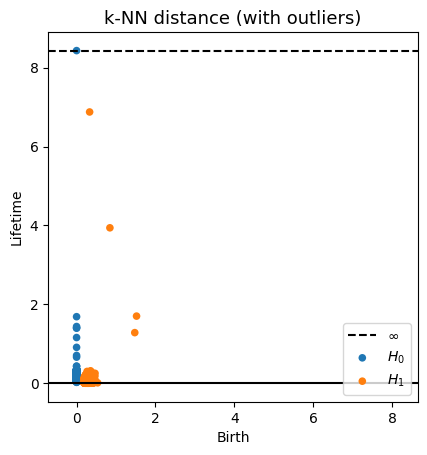
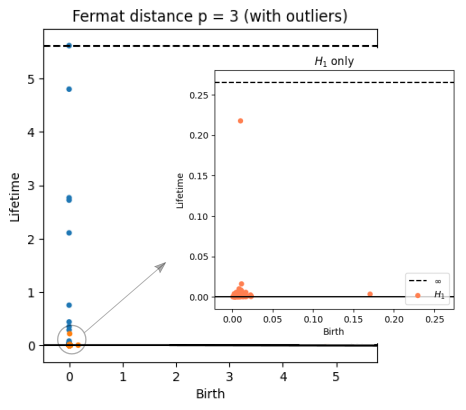
Fermat-based persistence diagrams
Robustness to outliers
Prop (F., Borghini, Mindlin & Groisman, 2023)
Let $\mathbb{X}_n$ be a sample of $\mathcal{M}$ and let $Y\subseteq \mathbb{R}^D\smallsetminus \mathcal{M}$ be a finite set of outliers.
Let $\delta = \displaystyle \min\Big\{\min_{y\in Y} d_E(y, Y\smallsetminus \{y\}), ~d_E(\mathbb X_n, Y)\Big\}$.
Then, for all $k>0$ and $p>1$,
\[
\mathrm{dgm}_k(\mathrm{Rips}_{<\delta^p}(\mathbb{X}_n \cup Y, d_{\mathbb{X}_n\cup Y, p})) = \mathrm{dgm}_k(\mathrm{Rips}_{<\delta^p}(\mathbb{X}_n, d_{\mathbb{X}_n, p}))
\]
where $\mathrm{Rips}_{<\delta^p}$ stands for the Rips filtration up to parameter $\delta^{p}$ and $\mathrm{dgm}_k$ for the persistent homology of deg $k$.

Fermat-based persistence diagrams
Computational implementation
- Complexity:
$O(n^3)$
reducible to $O(n^2*k*\log(n))$ using the $k$-NN-graph (for $k = O(\log n)$ the geodesics belong to the $k$-NN graph with high probability).
- Python library:
fermat
- Tool in Giotto-TDA:
In progress
- Computational experiments:
ximenafernandez/intrinsicPH
Applications to
time series analysis
$\bullet$ Fernandez X., Borghini E., Mindlin G. & Groisman P. Intrinsic persistent homology via density-based metric learning, Journal of Machine Learning Research, 2023 (to appear) arXiv:2012.07621
$\bullet$ Fernandez X., Mateos D. Topological biomarkers for real time detection of epileptic seizures, 2022. arXiv:2211.02523
Topological analysis of time series
- Signal:
$\varphi:\mathbb R \to \mathbb{R}$
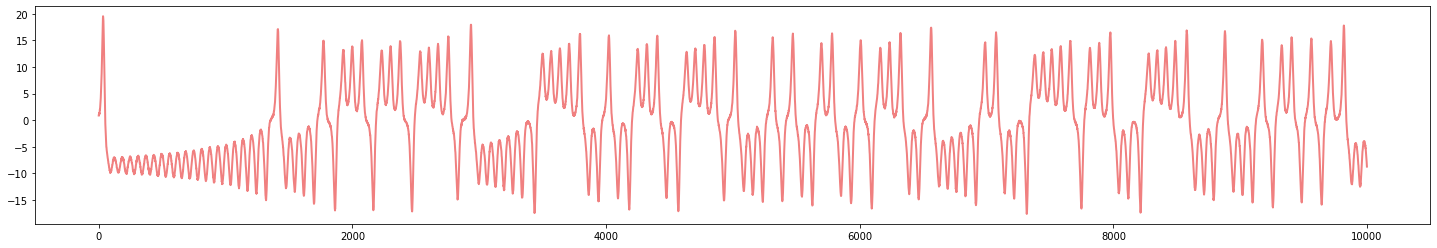
We assume that $\varphi$ is an observation of an underlying dynamical system $(\mathcal M, \phi)$, with $\mathcal M$ a topological space and $\phi\colon \mathbb R \times \mathcal M\to \mathcal M$ the evolution function.
That is, there exist an observation function $F:\mathcal M\to \mathbb R$ and an initial state $x_0\in \mathcal M$ such that \begin{align}\varphi = \varphi_{x_0}:\mathbb{R}&\to \mathbb{R}\\ t&\mapsto F(\phi_t(x_0)) \end{align}
Topological analysis of time series
- Signal:
$\varphi:\mathbb R \to \mathbb{R}$

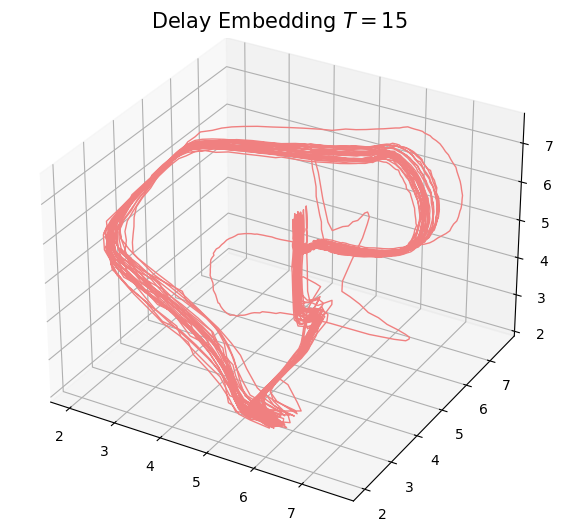
- Delay embedding: Given $T$ the time delay and $D$ the embedding dimension. \[\mathcal{M}_{T,D} = \{\big(\varphi(t), \varphi(t+T), \varphi(t+2 T) \dots, \varphi(t+(D-1)T)\big): t\in \mathbb R\}\subseteq \mathbb{R}^D\]
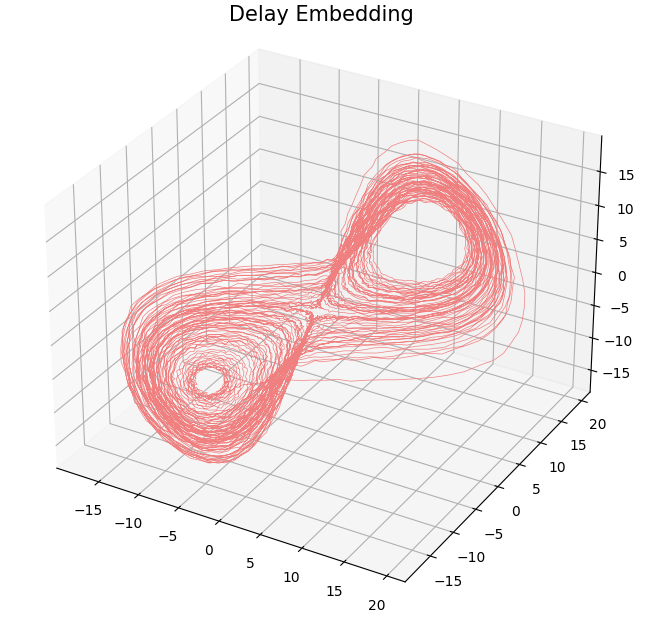
Topological analysis of time series
- Signal:
$\varphi:\mathbb R \to \mathbb{R}$

- Delay embedding: Given $T$ the time delay and $D$ the embedding dimension. \[\mathcal{M}_{T,D} = \{\big(\varphi(t), \varphi(t+T), \varphi(t+2 T) \dots, \varphi(t+(D-1)T)\big): t\in \mathbb R\}\subseteq \mathbb{R}^D\]
- Limit set: Given $(\mathcal M, \phi)$ a dynamical system and $x_0\in \mathcal M$, \[\mathcal A_{x_0} = \{x\in \mathcal M: \exists t_i\to \infty \text { with } \phi_{t_i}(x_0)\to x\}.\]

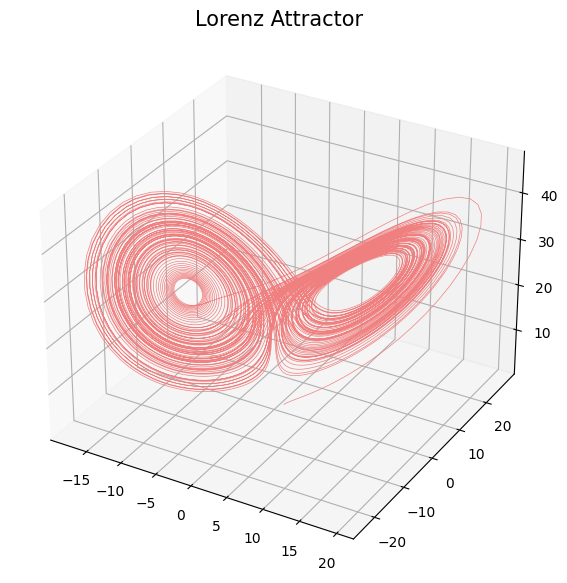
Topological analysis of time series
- Signal:
$\varphi:\mathbb R \to \mathbb{R}$

- Delay embedding: Given $T$ the time delay and $D$ the embedding dimension. \[\mathcal{M}_{T,D} (\phi) = \{\big(\varphi(t), \varphi(t+T), \varphi(t+2 T) \dots, \varphi(t+(D-1)T)\big): t\in \mathbb R\}\subseteq \mathbb{R}^D\]
- Limit set: Given $(\mathcal M, \phi)$ a dynamical system and $x_0\in \mathcal M$, \[\mathcal A_{x_0} = \{x\in \mathcal M: \exists t_i\to \infty \text { with } \phi_{t_i}(x_0)\to x\}.\]
- Theorem (Takens).* Let $\mathcal{M}$ be a smooth, compact, Riemannian manifold. Let $T> 0$ be a real number and let $D > 2 \mathrm{dim}(\mathcal{M})$ be an integer. Then, for generic $\phi \in C^2(\mathbb{R} \times \mathcal{M}, \mathcal{M})$, $F\in C^2(\mathcal{M}, \mathbb{R})$ and $x_0\in \mathcal M$, if $\varphi_{x_0} = F(\phi_\bullet(x_0))$ is an observation of $(\mathcal M, \phi)$, then the limit set $\mathcal A_{x_0}$ is 'diffeomorphic'$^{**}$ to $\mathcal{M}_{T,D} (\varphi_{x_0})$.
** There exists $\psi:\mathcal M\to \mathbb R^{D}$ an embedding such that $\psi|_{\mathcal A_{x_0}}: \mathcal A_{x_0}\to \mathcal{M}_{T,D} (\varphi_{x_0})$ is a bijection.
*Corollary 5, Detecting strange attractors in tubulence, F. Takens, 1971.
Parameter selection
Embedding dimension $D$
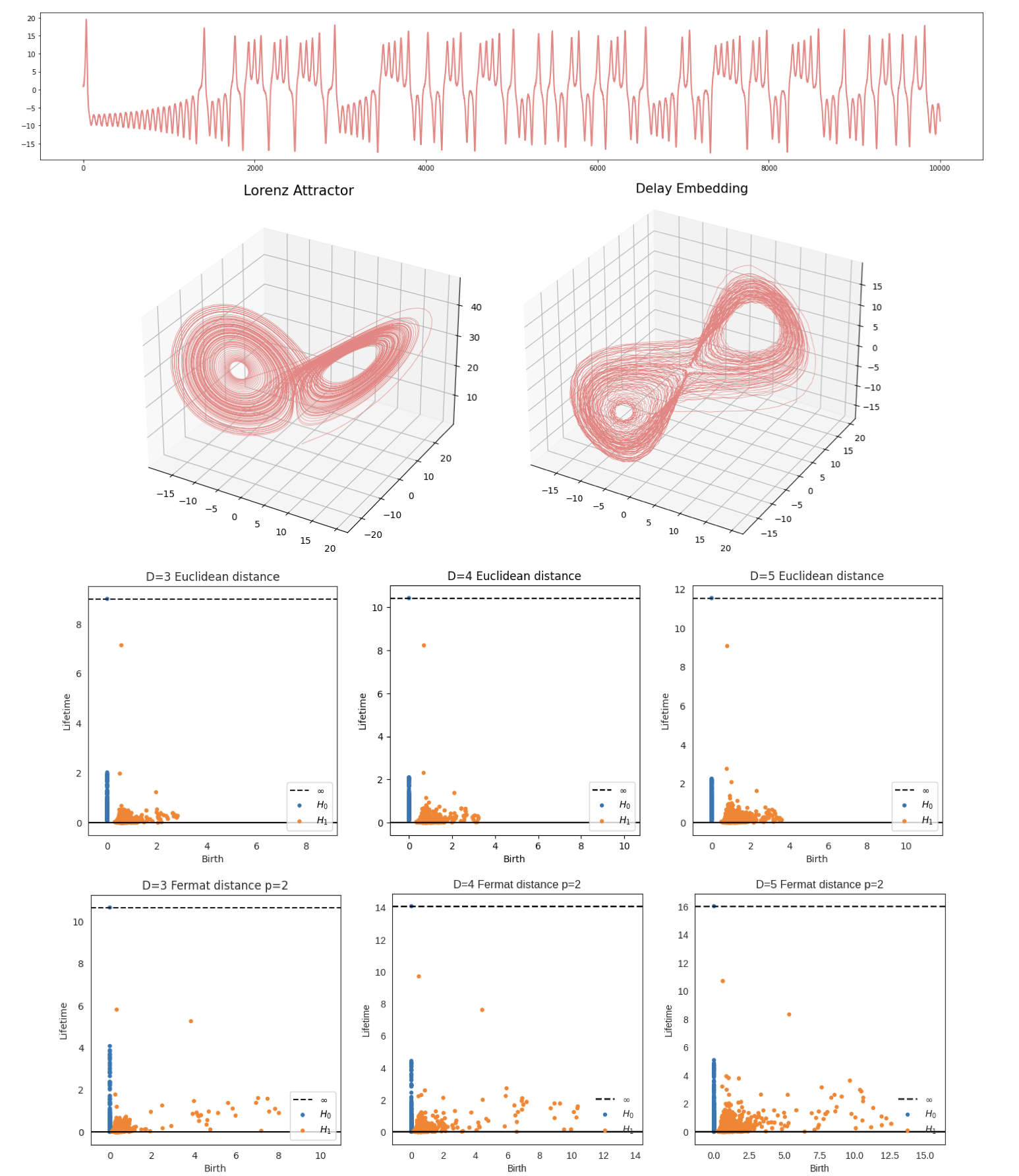
Parameter selection
Time delay $T$
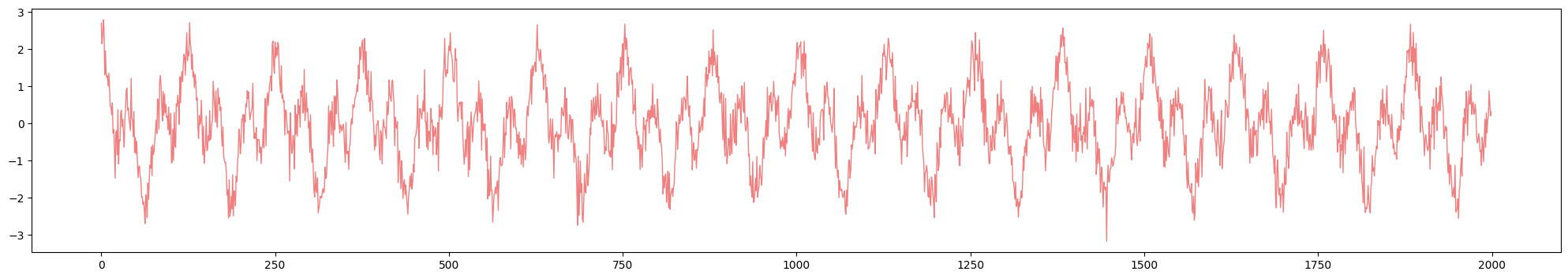
Anomaly detection
Electrocardiogram
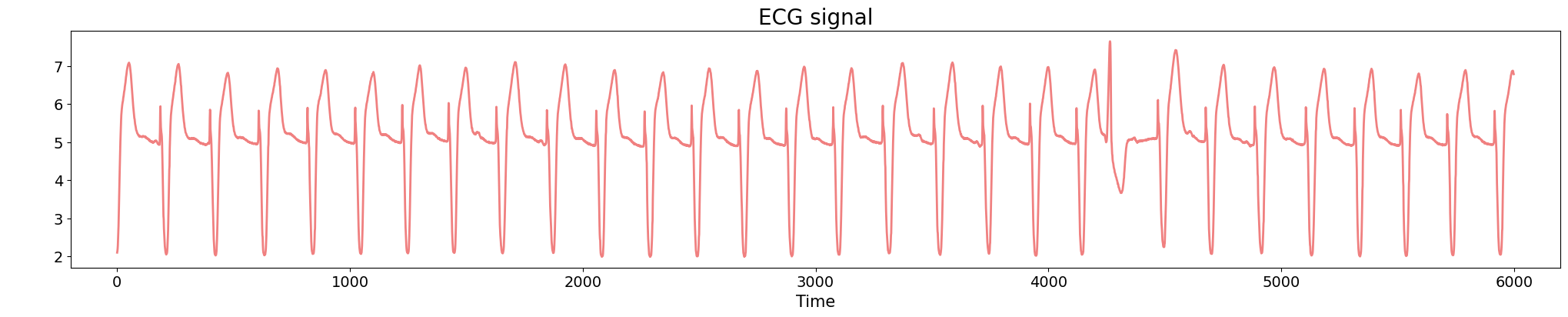
Source data: PhysioNet Database https://physionet.org/about/database/
Anomaly detection
Electrocardiogram

Anomaly detection
Electrocardiogram
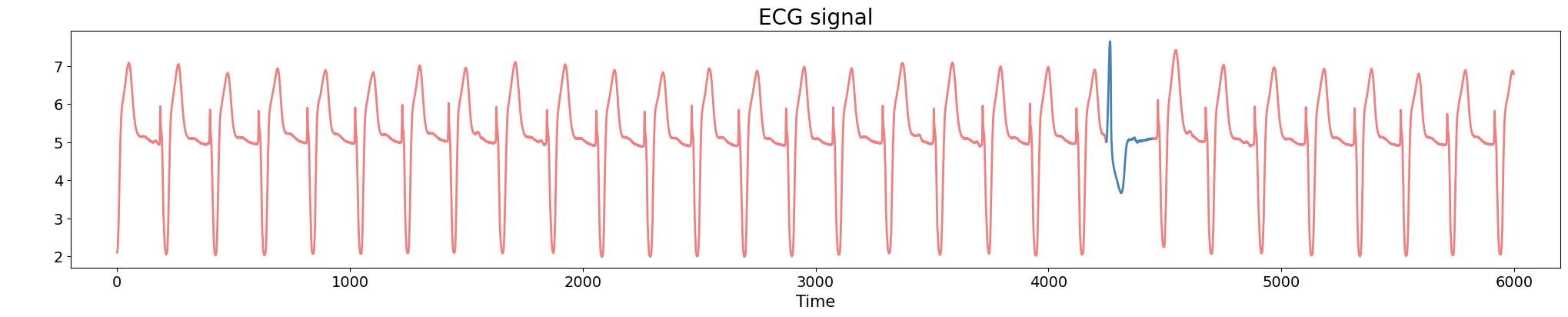
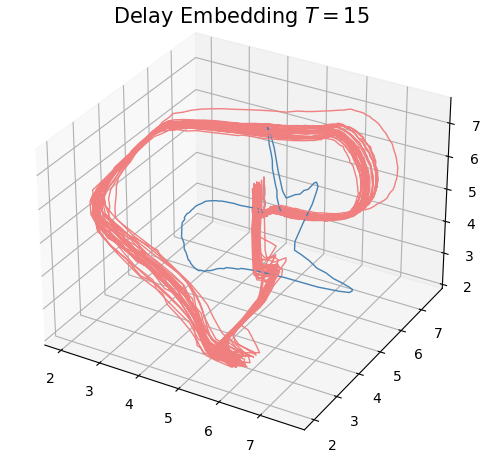
Anomaly detection
Electrocardiogram


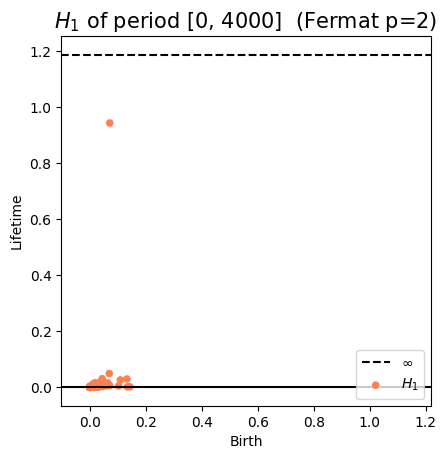
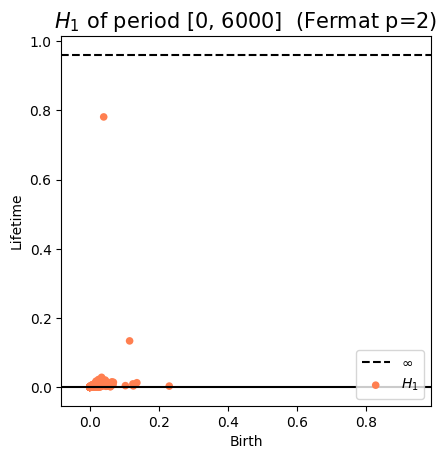
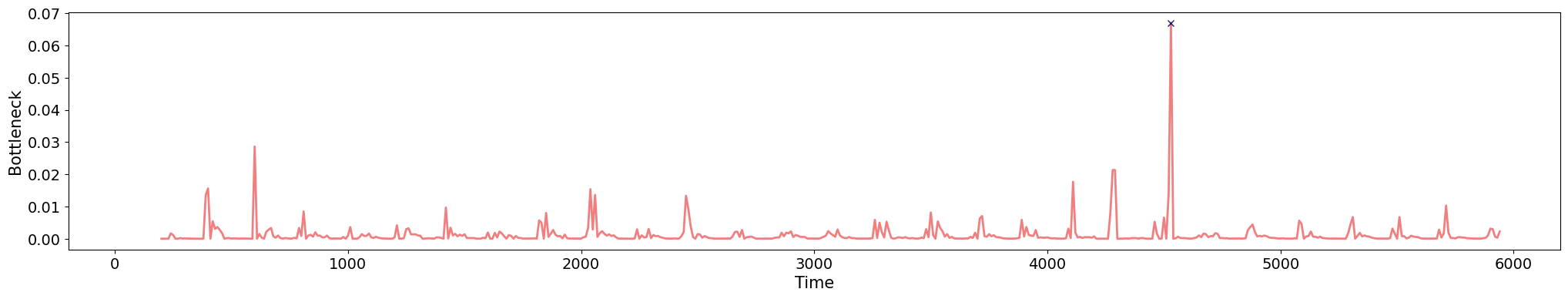
Anomaly detection
Electrocardiogram
 \[t\mapsto \mathrm{dgm_1}(\mathrm{Rips}(\mathcal M_{T, D}\varphi|_{[0,t]}))\]
\[t\mapsto \mathrm{dgm_1}(\mathrm{Rips}(\mathcal M_{T, D}\varphi|_{[0,t]}))\]
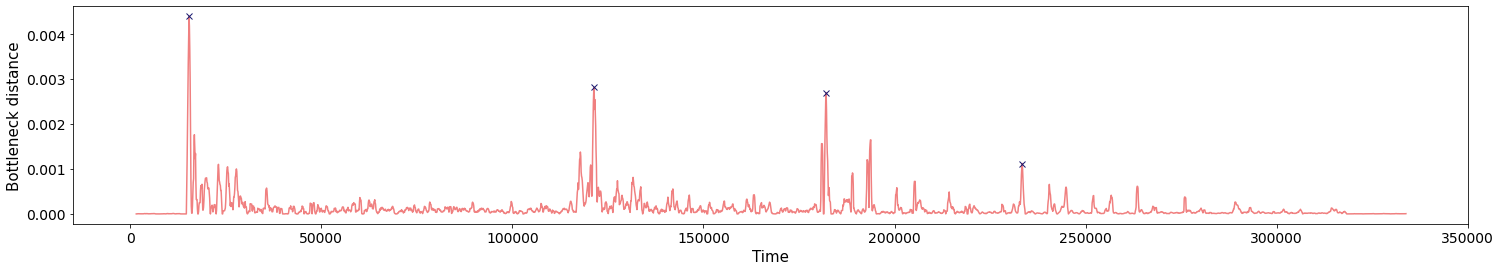
Change-points detection
Birdsongs
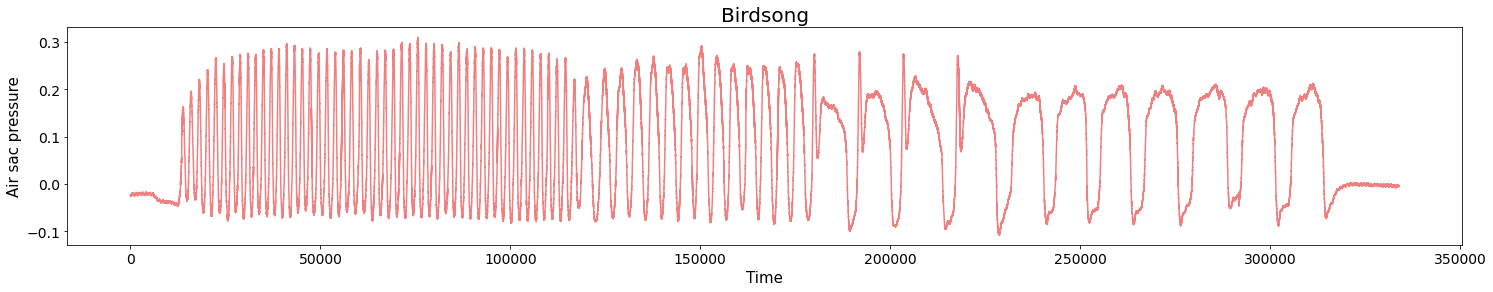
Source data: Private experiments. Laboratory of Dynamical Systems, University of Buenos Aires.
Change-points detection
Birdsongs

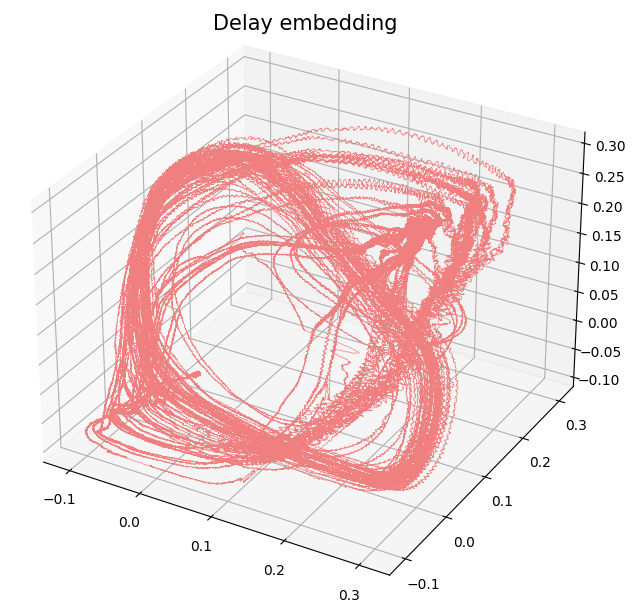
Change-points detection
Birdsongs


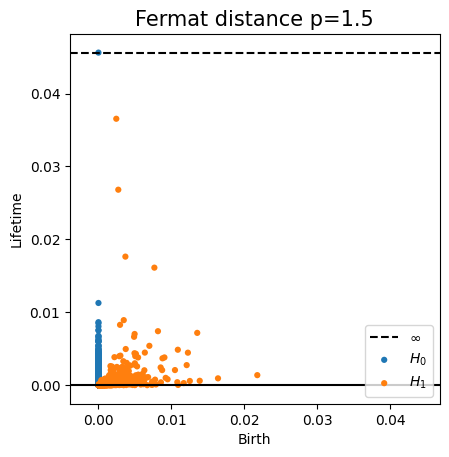
Change-points detection
Birdsongs
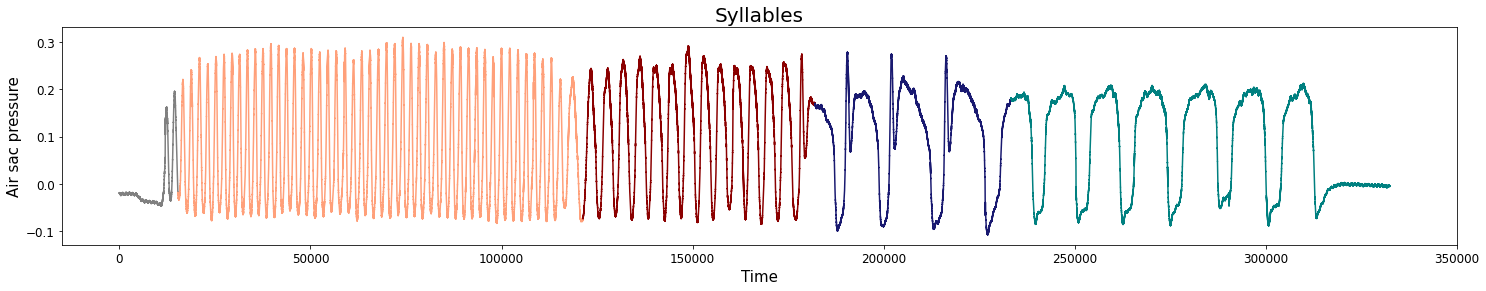
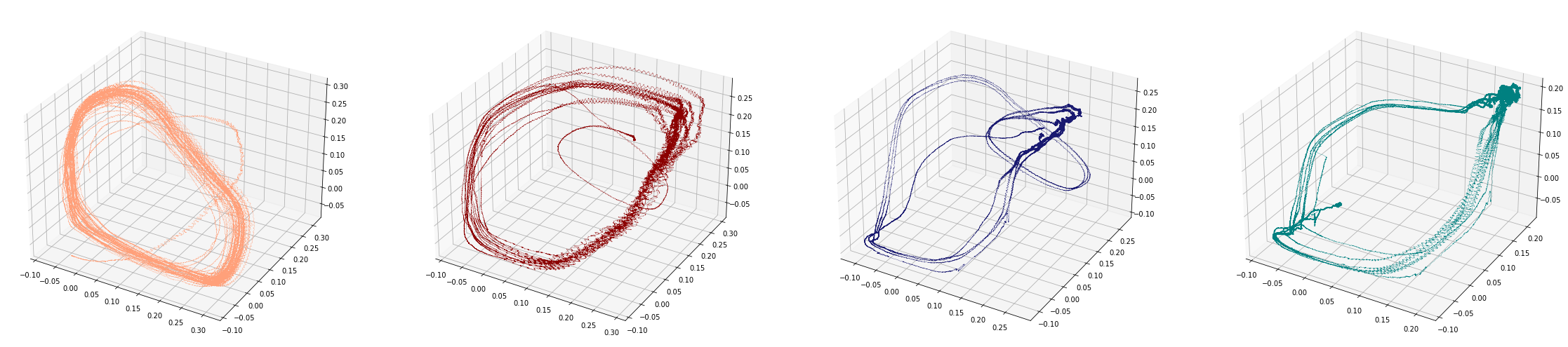

Change-points detection
Birdsongs
 \[t\mapsto \mathrm{dgm_1}(\mathrm{Rips}(\mathcal M_{T, D}\varphi|_{[0,t]}))\]
\[t\mapsto \mathrm{dgm_1}(\mathrm{Rips}(\mathcal M_{T, D}\varphi|_{[0,t]}))\]
Epileptic seizure detection
Epileptic seizure detection
EEG

Source: Open database CHB-MIT, Massachusetts Institute of Technology, USA.
Epileptic seizure detection
EEG

Source: Open database CHB-MIT, Massachusetts Institute of Technology, USA.
Epileptic seizure detection
iEEG
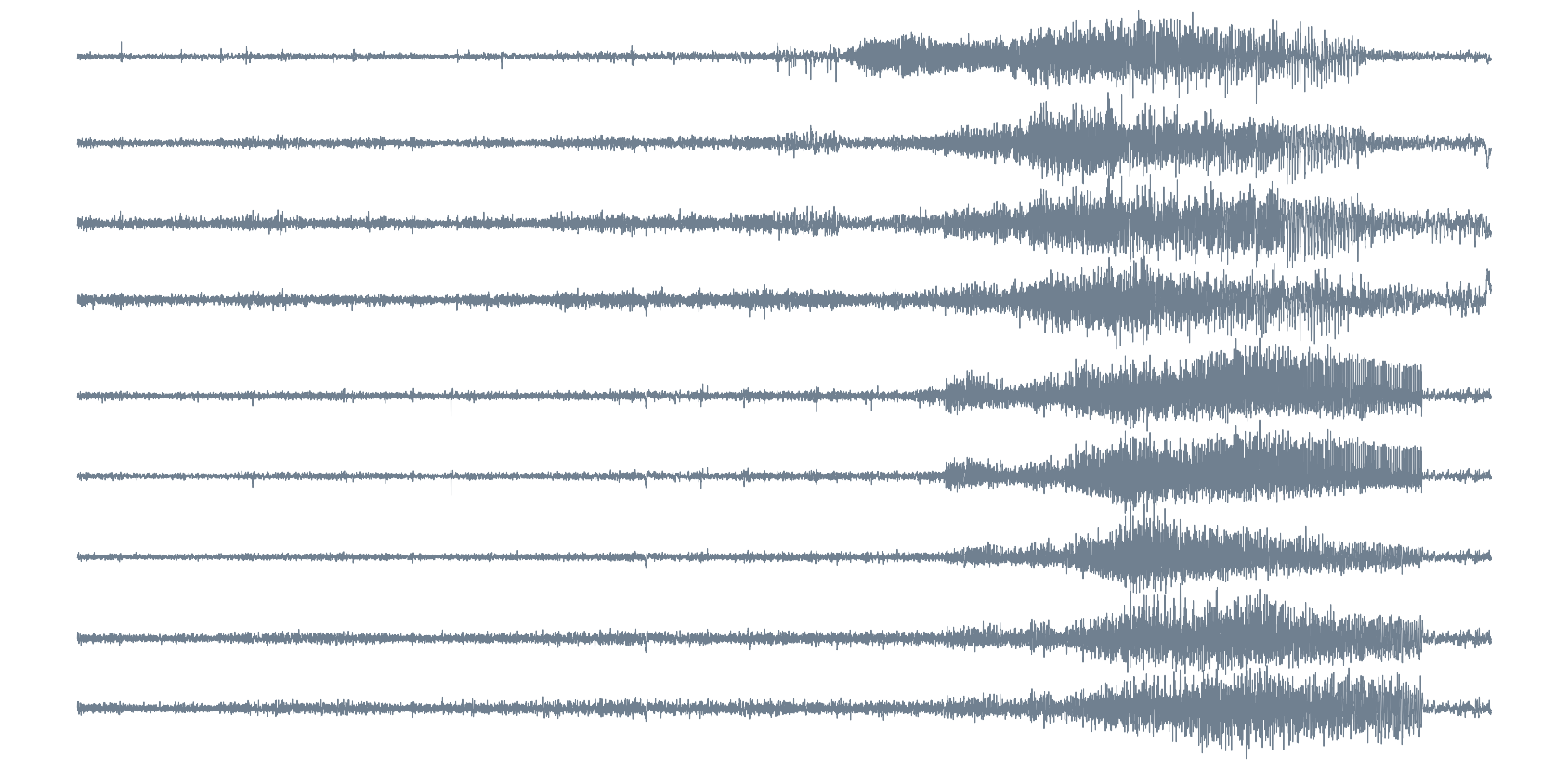
Source: Private recording, Toronto Western Hospital, Canada.
Epileptic seizure detection
iEEG

Source: Private recording, Toronto Western Hospital, Canada.
Standard signal analysis

Standard signal analysis
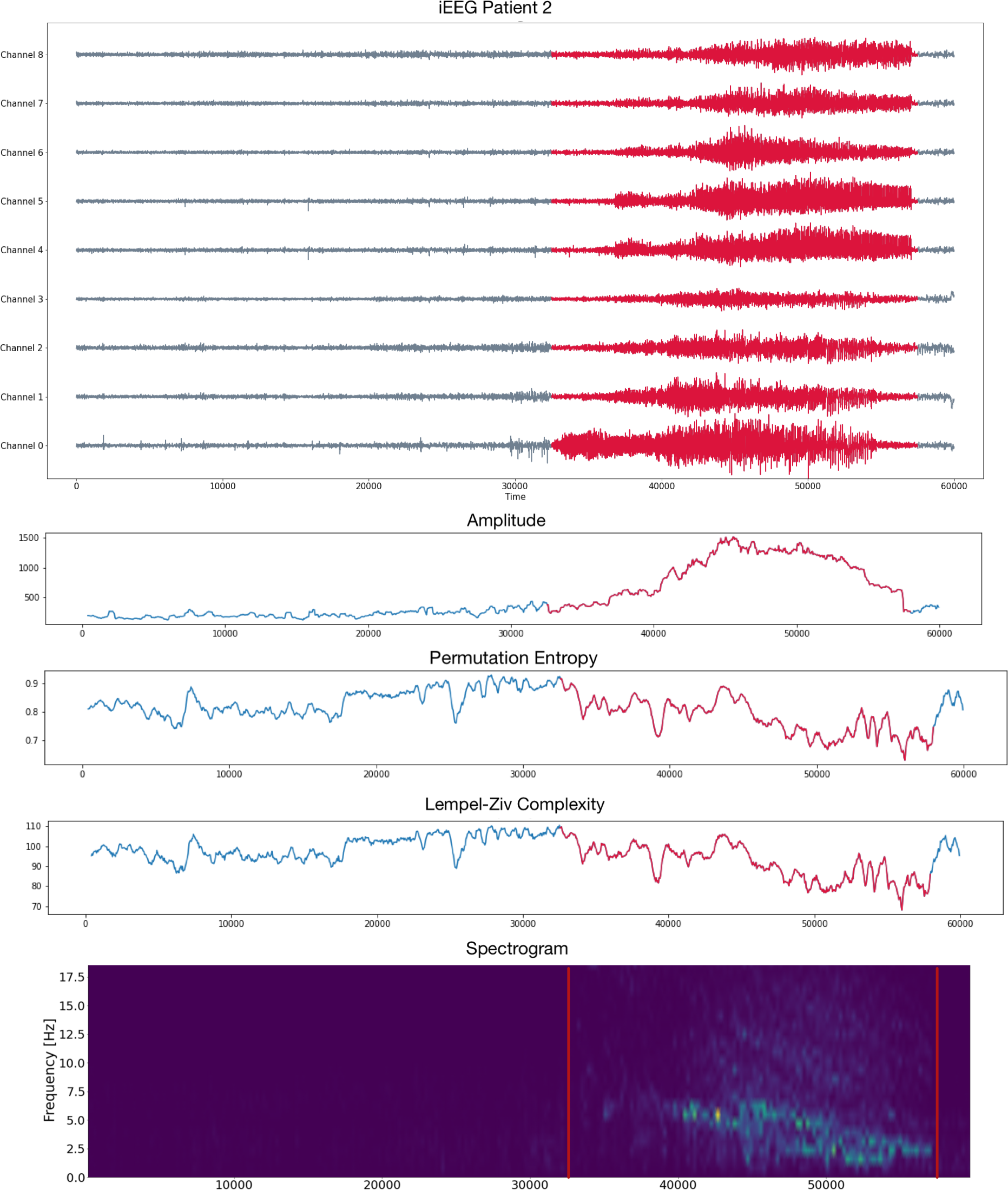
Brain Dynamics and Epilepsy
- Let $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ be a multichannel recording.$~~~~~~~~~~~~~~~~~~$$~~~~~~~~~~~~~~~~$$~~~~~~$
Brain Dynamics and Epilepsy
- Let $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ be a multichannel recording.$~~~~~~~~~~~~~~~~~~$$~~~~~~~~~~~~~~~~$$~~~~~~$
- We assume there exist an underlying dynamical system $(X, \phi)$ such that the time series $\{\varphi_i\}_{1\leq i \leq n}$ are the result of observation functions $\{F_i:X\to \mathbb R\}_{1\leq i \leq n}$
(i.e. $\varphi_i(t) = F_i(\phi(t,x))$ for some initial state $x\in X$).
Brain Dynamics and Epilepsy
- Let $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ be a multichannel recording.$~~~~~~~~~~~~~~~~~~$$~~~~~~~~~~~~~~~~$$~~~~~~$
- We assume there exist an underlying dynamical system $(X, \phi)$ such that the time series $\{\varphi_i\}_{1\leq i \leq n}$ are the result of observation functions $\{F_i:X\to \mathbb R\}_{1\leq i \leq n}$
(i.e. $\varphi_i(t) = F_i(\phi(t,x))$ for some initial state $x\in X$). - We analyze the underlying (unknown) dynamics by estimating their attractors (that is, the asymptotic states of the system) via the simultaneous embedding of the signals.
Brain Dynamics and Epilepsy
- Let $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ be a multichannel recording.$~~~~~~~~~~~~~~~~~~$$~~~~~~~~~~~~~~~~$$~~~~~~$
- We assume there exist an underlying dynamical system $(X, \phi)$ such that the time series $\{\varphi_i\}_{1\leq i \leq n}$ are the result of observation functions $\{F_i:X\to \mathbb R\}_{1\leq i \leq n}$
(i.e. $\varphi_i(t) = F_i(\phi(t,x))$ for some initial state $x\in X$). - We analyze the underlying (unknown) dynamics by estimating their attractors (that is, the asymptotic states of the system) via the simultaneous embedding of the signals.
Topological biomarkers
- Let $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ be a multichannel recording.$~~~~~~~~~~~~~~~~~~$$~~~~~~~~~~~~~~~~$$~~~~~~$
- Given a window size $\omega$, we compute for every $t\in \mathbb R$ the embedding $\mathrm{SWE}_{\varphi, \omega}(t) :=\varphi([t-\omega, t])$ in $\mathbb{R}^n$.
Topological biomarkers
- Let $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ be a multichannel
recording.$~~~~~~~~~~~~~~~~~~$$~~~~~~~~~~~~~~~~$$~~~~~~$
- Given a window size $\omega$, we compute for every $t\in \mathbb R$ the embedding $\mathrm{SWE}_{\varphi, \omega}(t) :=\varphi([t-\omega, t])$ in $\mathbb{R}^n$.
- We compute the persistent homology $\mathrm{dgm}(\mathrm{SWE}_{\varphi, \omega}(t))$ of the time evolving sliding window embedding.

Topological biomarkers
- Let $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ be a multichannel recording.$~~~~~~~~~~~~~~~~~~$$~~~~~~~~~~~~~~~~$$~~~~~~$
- Given a window size $\omega$, we compute for every $t\in \mathbb R$ the embedding $\mathrm{SWE}_{\varphi, \omega}(t) :=\varphi([t-\omega, t])$ in $\mathbb{R}^n$.
- We compute the persistent homology $\mathrm{dgm}(\mathrm{SWE}_{\varphi, \omega}(t))$ of the time evolving sliding window embedding.
Topological biomarkers
Given $\varphi = (\varphi_1, \varphi_2, \dots \varphi_n):\mathbb R\to \mathbb{R}^n$ a multichannel recording and $\omega$ a window size.
\[\mathbb{R} ~\longrightarrow ~\mathrm{Top} ~\longrightarrow ~\mathrm{Dgm}~~~~~~~~~~~~~~~~~~~~~~~~~~~~\] \[t\mapsto \mathrm{SWE}_{\varphi, \omega} (t)\mapsto \mathrm{dgm}\big(\mathrm{SWE}_{\varphi, \omega}(t) \big)=:\mathcal{D}_t\]
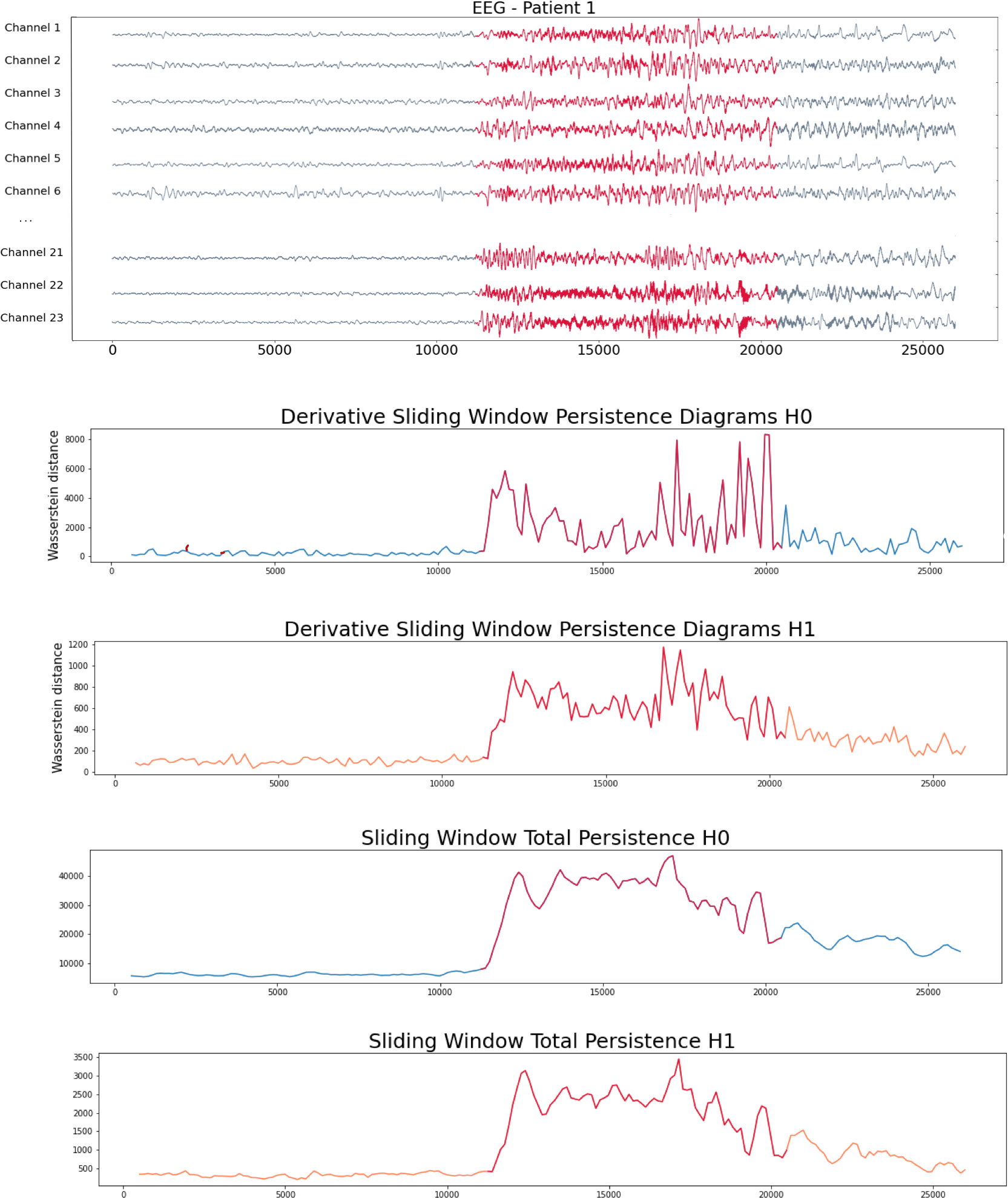
We compute the following topological summaries:
- Approximate Derivative: Given a time step $\varepsilon>0$\[\dfrac{d_{W}(\mathcal{D}_t, \mathcal{D}_{t-\varepsilon})}{\varepsilon}\]
- Total persistence: \[\sum_{(x,y) \in \mathcal{D_t}} \mathrm{pers}(x,y).\]
Patient 1 (EEG)
- Scalp EEG with 23 sensors at frequency 256 Hz.
- Source: CHB-MIT database from Children’s Hospital Boston, USA.
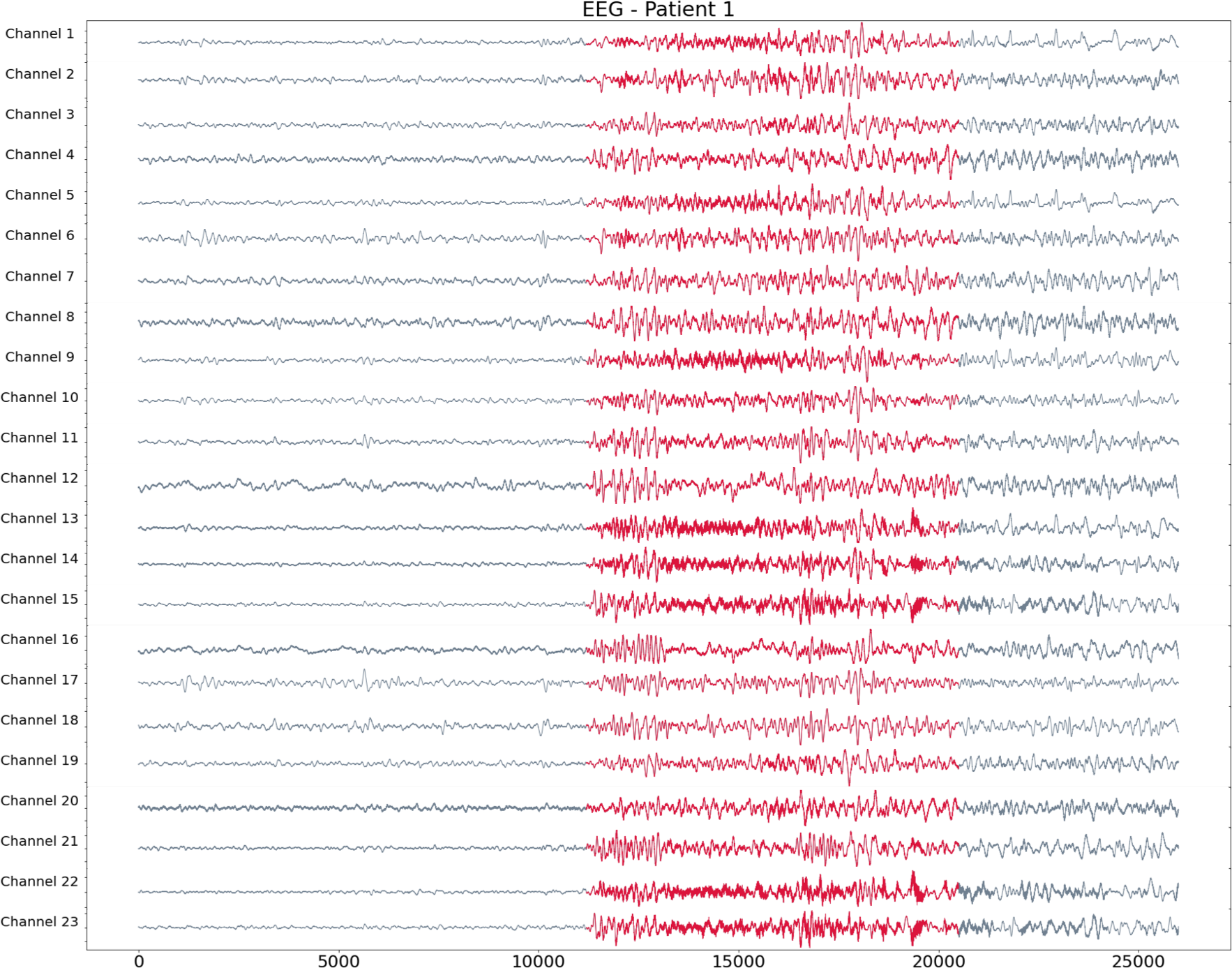
Patient 1 (EEG)
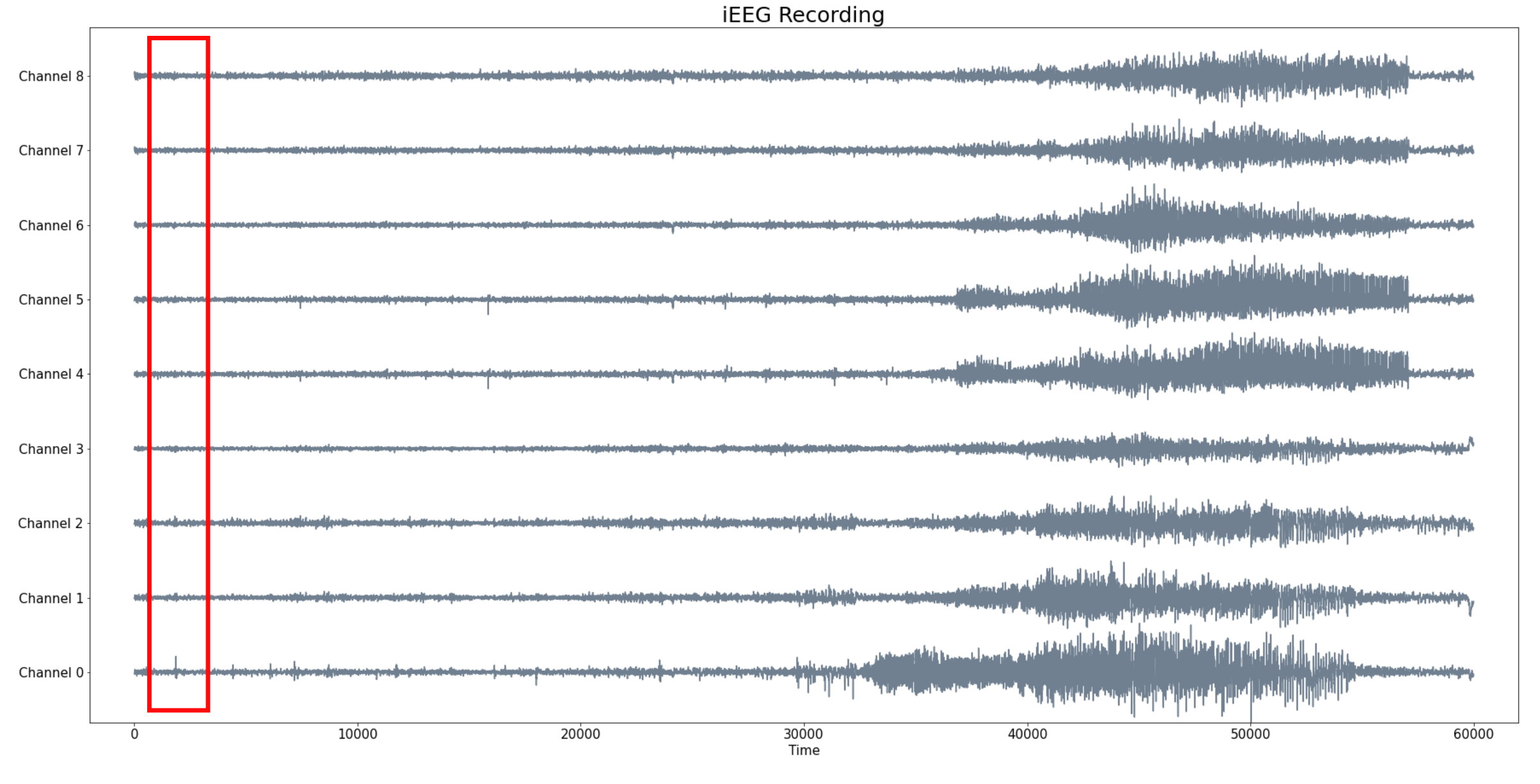
Patient 2 (iEEG)
- Intracraneal EEG with 9 sensors at frequency 200 Hz.
- Source: Clinical presurgical analysis, Toronto Western Hospital, Canada.
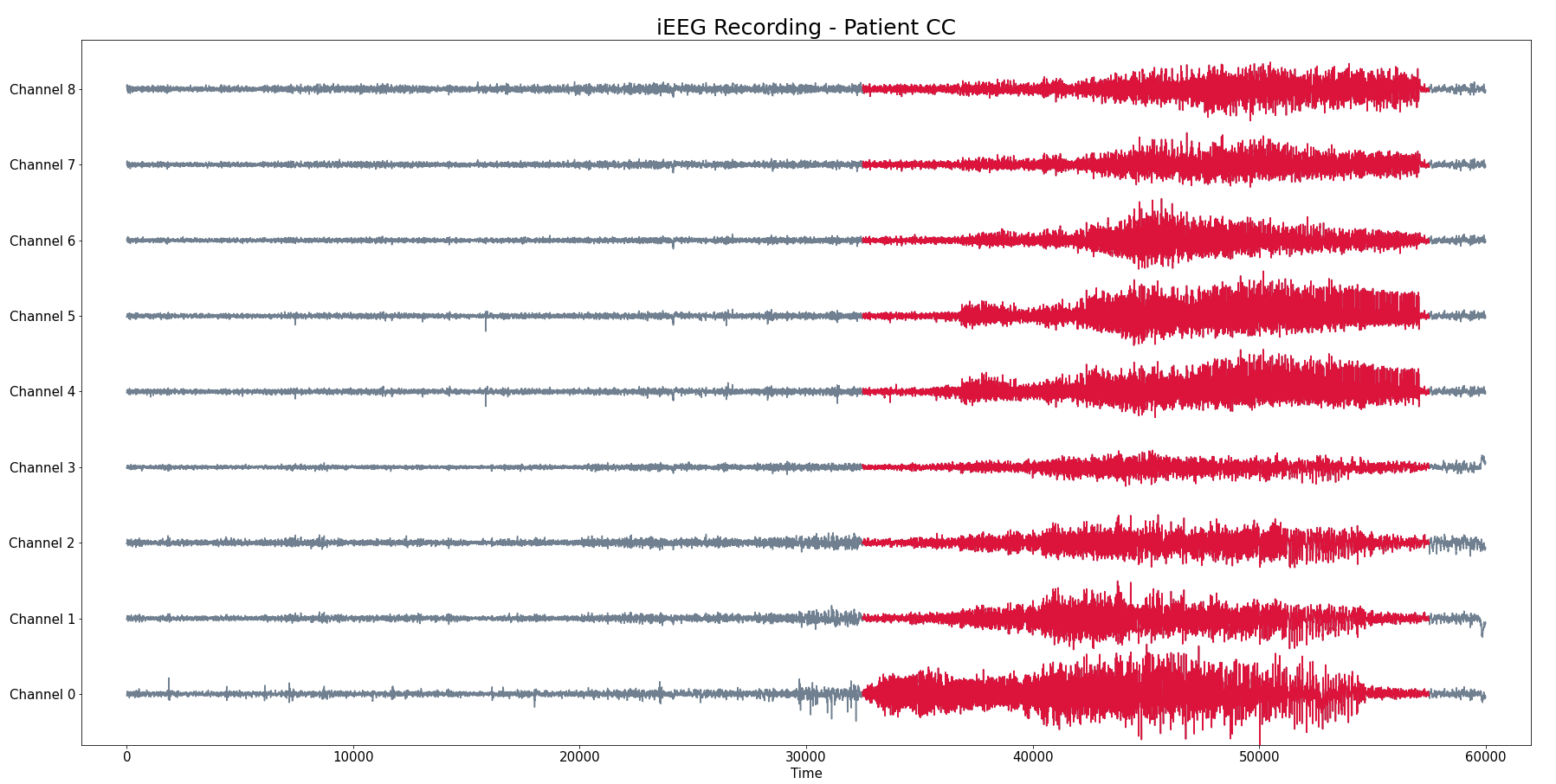
Patient 2 (iEEG)
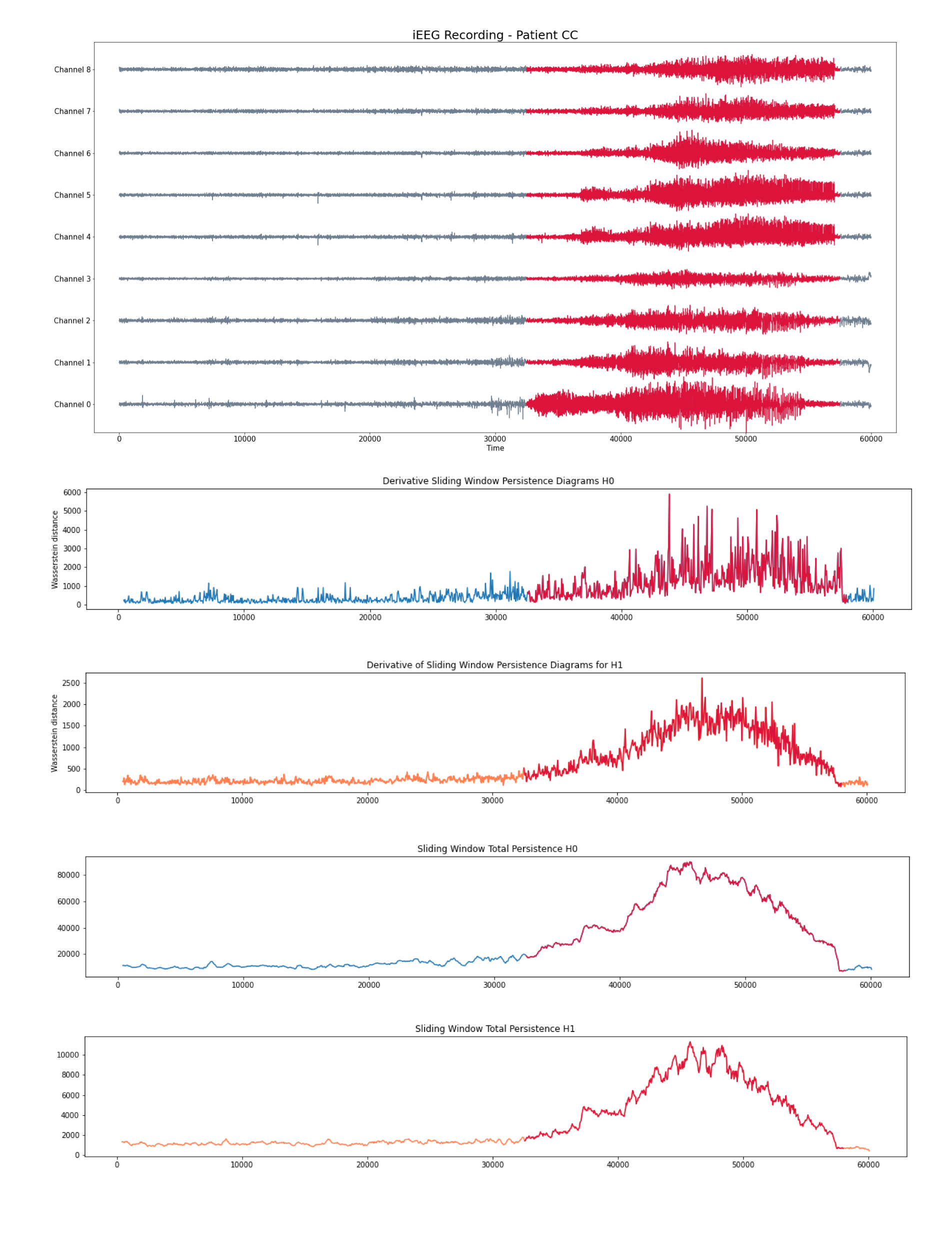
Analysis by channel
Given $\varphi_i:\mathbb R\to \mathbb{R}$ a single channel of the recording, we construct its time delay embedding $\mathcal M_{T, D}\varphi_i$.
Given $\omega$ a window size, the sliding window delay embedding $\mathrm{SWDE}_{\varphi_i, \omega}$ of $\varphi_i$ is the sliding window embedding of $\mathcal M_{T, D}\varphi_i$.
\[~~~~~\mathbb R ~\longrightarrow ~~\mathrm{Top} ~~\longrightarrow ~~\mathrm{Dgm}~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\] \[~~~~~~~~~~~~t\mapsto \mathrm{SWDE}_{\varphi_i, \omega} (t)\mapsto \mathrm{dgm}\big(\mathrm{SWDE}_{\varphi_i, \omega}(t) \big)=:\mathcal{D}_t\]
Analysis by channel


References
- Density-based intrinsic persistent homology
- Source: X. F., E. Borghini, G. Mindlin, P. Groisman, Intrinsic persistent homology via density-based metric learning. Journal of Machine Learning Research (to appear), 2023. ArXiv:2012.07621 (2020)
- Github Repository: ximenafernandez/intrinsicPH
- Tutorial: Intrinsic persistent homology. AATRN Youtube Channel (2021)
- Epilepsy
- Source: X. F., D. Mateos, Topological biomarkers for real time detection of epileptic seizures. arXiv:2211.02523 (2022)
- Github Repository: ximenafernandez/epilepsy
github.com/ximenafernandez $~~~~$ ximenafernandez.github.io/ $~~~~$ ximena.l.fernandez@durham.ac.uk $~~~~$ @pi_ene